By your requests we implemented new style home pages, for your convenience. The new portal should look and feel what many of you called Outlook style or Explorer style: project based folders you create on the left, lists of objects on the top right, and the content on the bottom right.
The older home page interface is temporarily available by clicking this notification message. As soon as we migrate all of the tools to the new interface, we make sure that you like it and it sticks, the older home page will be depreciated and discontinued.
We are aware that just like anything else new, some issues may come up, some problems may appear while using the new system. We are working hard to fix those ! But we wanted you to know that we invite your comments. Please, report ANY problems you see. We know that you, our users play an important and critical role in creating HIVE, we appreciate every step, any recommendation, an inquiry and a desire to make HIVE better.
When you talk, we listen!
Please, use the new Home Pages! Enjoy the new interface!.
",,?cmd=oldhomeDrugVar is an integrated database of germline and somatic non-synonymous single nucleotide variation associated with drug binding. By integrating germline and somatic nsSNVs (non-synonymous single nucleotide variations) from a variety of source and protein-drug structure information from PDB (Protein Data Bank), 13011 protein-drug binding sites and 3133 nsSNVs have been identified within interaction between 253 proteins and 235 drugs.
Protein-drug structure information is retrieved from Anatomical Therapeutic Chemical Classification Browser.
Germline nsSNVs are retrieved from dbSNP.
Somatic nsSNVs are retrieved from TCGA, ICGA, IntOGen, COSMIC, UniprotKB and ClinVar.
Protein-drug structure information is manually filtered to exclude non-drug entries (Proteins, ions and metal) parsed by a program called OS to identified amino acid-drug binding site (distance within 4.5 Å)
All protein-drug binding sites are mapped to nsSNVs dataset to identify nsSNVs that affect protein-drug binding.
DrugVar can be searched by multiple modes: gene (UniProt AC, PDB ID and Gene name) or drug (Drug name, Drugbank ID, CAS ID and CID ID). Do not forget to specify the query type when using a specific identifier (searching by PDB ID when query type is set as other than PDB ID will show no results). Results page shows two sections: Protein-drug binding and nsSNVs that affect protein-drug binding.
For a comprehensive view, protein and drug identifiers are linked to different databases.
UniProtKB AC links to the UniProt database where the amino acid binding position and reference is highlighted.
PDB ID links to the PDB structure webset of the exact PDB identifier. Users can check the 3D view of amino acid-drug binding structure and other structure related information (publication, molecular description, structure validation and et). Users can also draw or zoom the figure to get the better view of the 3D structure.
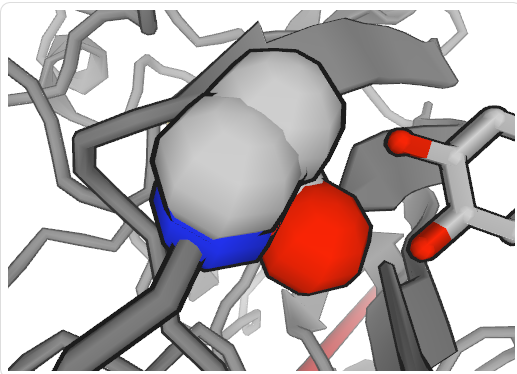
Figure shows binding between 4A7V (SOD1) and Dopamine at PDB position 28.
Atoms are colored by element:
Carbon - green
Hydrogen - white
Oxygen - red
Nitrogen - blue
Sulfur - yellow
Phosphorus - orange
Ligand ID links to the ligand description page in PDB where users can find ligand structure related information.
CID ID links to PubChem database which provides chemical information about the drug of interest.
At the DrugVar home page, users can identify protein-drug binding affecting nsSNVs in their own data using the scan function. The input file must be tab-delimited tsv or txt format and contains RefSeq accession, amino acid position and the wild type amino acid position (other information is optional). Users also need to specify the columns that contain the mandatory information and if their input file contains header. After submitting an input file, the results will be automatically downloaded as a tab deliminated tsv formatted file. For identified nsSNVs based on the input information, the original information will remain with the identified nsSNVs appended at the end.
In addition, users can scan mutations by specifying UniProt Accession and Protein Position. The output file will returns list of mutations available in DrugVar. if amino acid position of specified protein does not contain mutations in DrugVar, a superlink will provided linking to BioMuta, a curated single-nucleotide variation (SNV) and disease association database where the variations are mapped to the genome/protein/gene, where users can further check the if any mutations exist.
Each search result can be downloaded as a CSV file by clicking the download button. Users can also download both entire protein-drug binding sites dataset and protein-drug binding affecting nsSNVs sites dataset by clicking download button on the DrugVar home page.